Modular risk analysis to assess complex risk pathways: An application to farm biosecurity
Natalia Ciria1, Giovanna Ciaravino1, Alberto Allepuz1
1Universitat Autònoma de Barcelona, Spain
1. Background
Quantitative risk analysis, using Monte Carlo simulations, is a method to numerically determine the probability of a risk event occurring taking into account uncertainty. It considers all possible values that each variable could take and the probability of their occurrence.
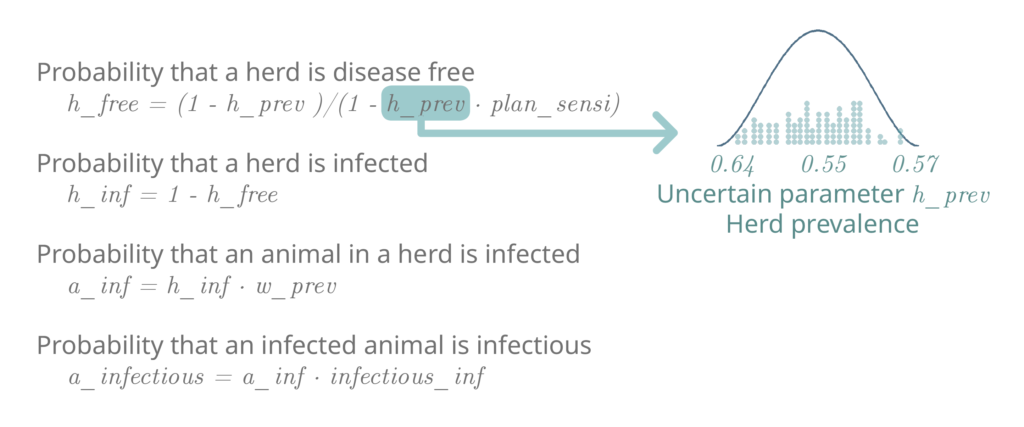
For example: To calculate the probability of buying an infectious animal from a farm, we use surveillance estimates that indicate the herd prevalence of BVD in the farm region is between 0.64 and 0.57, with a mean of 0.55.
2. Problem
Detailed risk quantification in complex systems, such as the different pathways by which pathogens enter a farm, involves numerous steps and parameters that make the model unwieldy and difficult to analyse, explain and visualise.
3. Methods
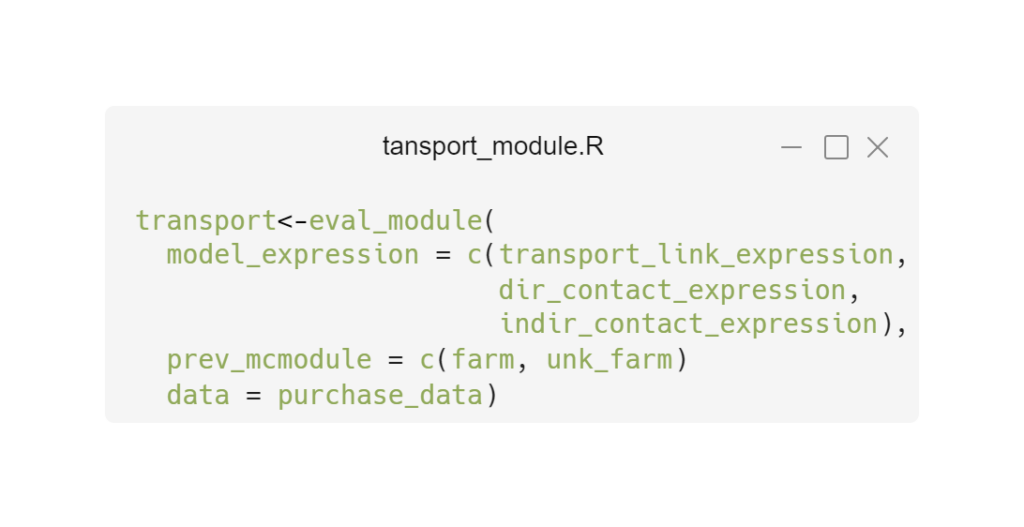
Modular risk analysis helps to analyse complex systems by looking at parts of a system separately and then combining the results for an overall assessment. Our modular risk analysis approach consists of mathematical expressions grouped into modules.
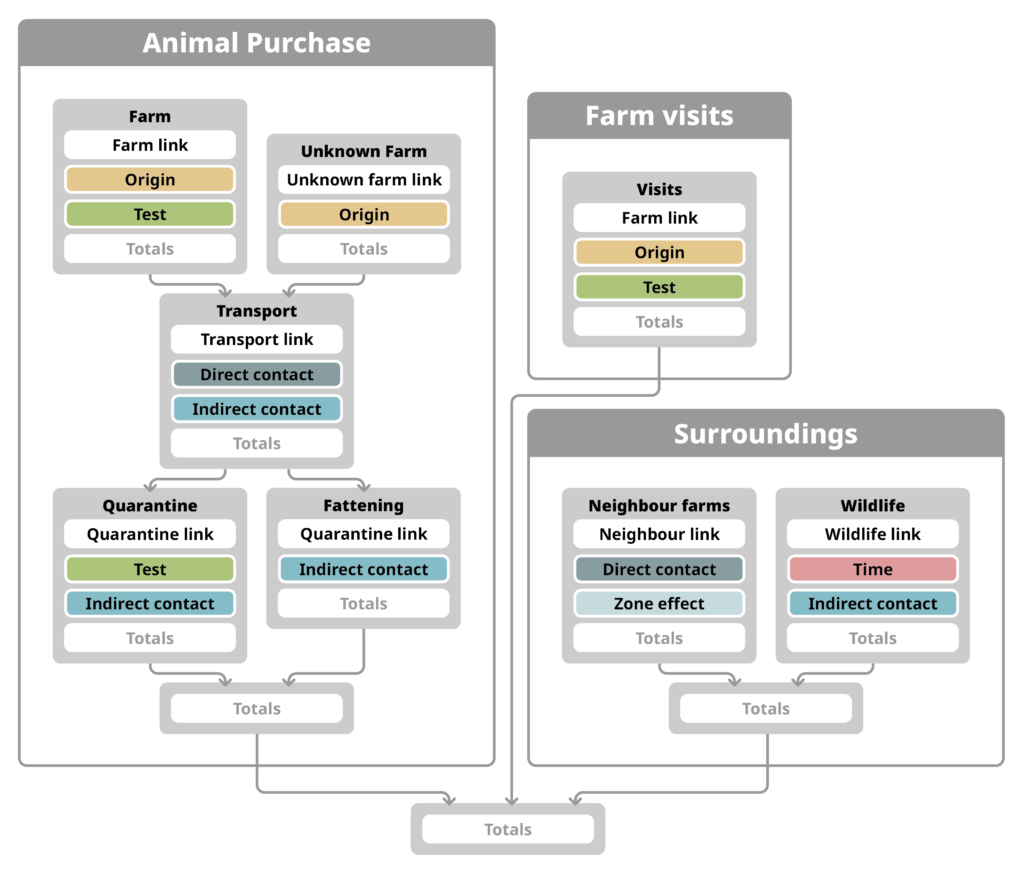
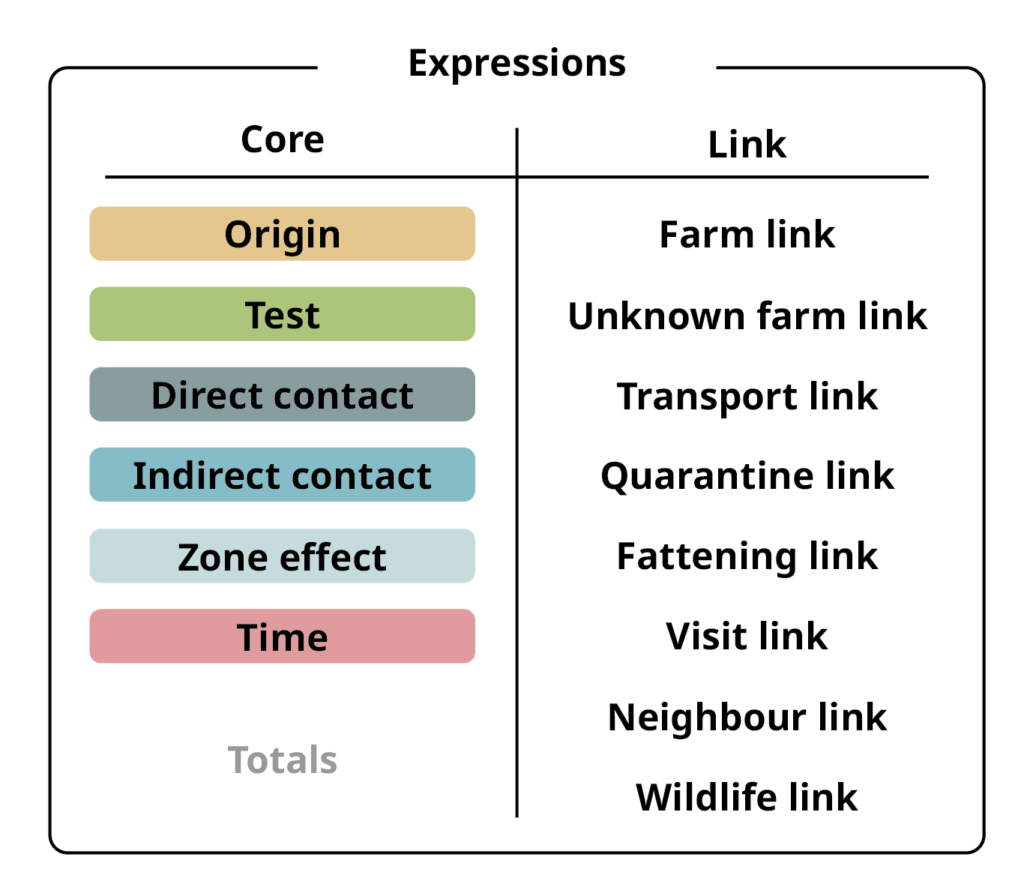
Expressions: Set of mathematical functions. Core expressions calculate probabilities, link expressions connect core expressions with different inputs and between modules.
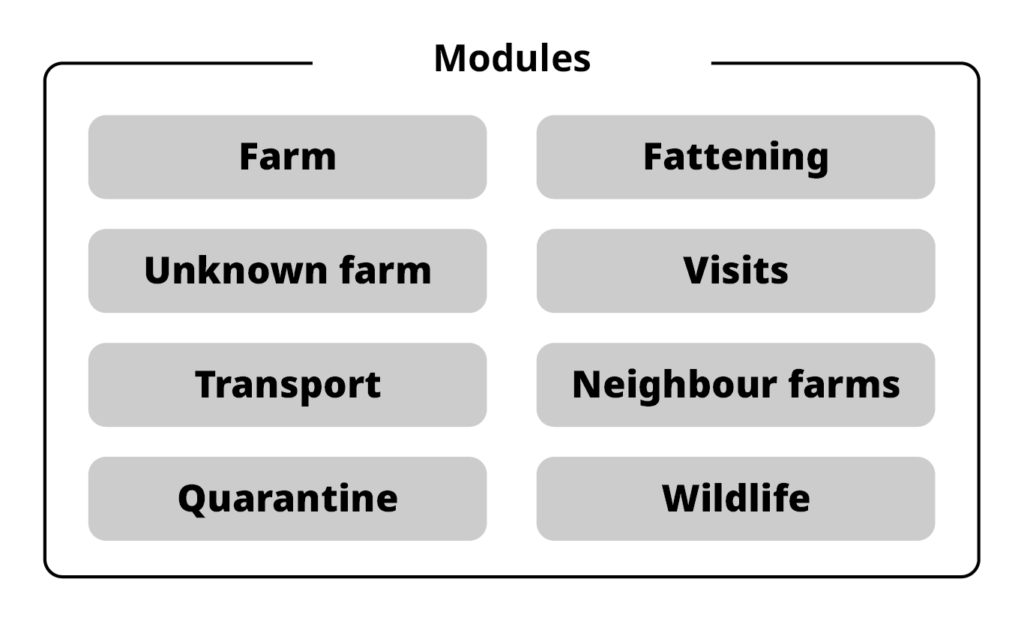
Modules: Set of expressions and input data that constitute a self-contained part of the system.
4. Implementation in R
Based on the mc2d package, we are developing a new package to automate the creation of stochastic variables, simplify the building of modular models and visualise the model and its results.
5. Next steps
We are testing this model on real cattle farms to assess the probability of pathogen introduction (IBR, BVD and TB) and the impact of biosecurity measures, and will extend it to other species and pathogens.
We plan to publish the model’s framework as an R package that could be applied to other types of risk analysis beyond biosecurity.
Contact: natalia.ciria@uab.cat


This research project was funded by BIOSECURE Horizon Europe project (www.biosecure.eu)